Projects
Active Projects
Genomics-based Technologies in Forest Tree Breeding Biotechnology and bioinformatics offer a wide spectrum of possibilities to decode, study and propose models based on tree genome sequences. The improvement and application of new molecular and genetic tools could sharply reduce the time required to grow the trees and help to identify trees with certain characteristics that confer resistance to pest infestation and environmental changes.
Next Generation genome annotation
Information coming soon.
Genotyping for balsam woolly adelgid tolerance
Information coming soon.
NGS technologies in tree improvement and conservation genetics of Dipteryx oleifera Benth.
Information coming soon.
Post-harvest Needle Abscission in Fir species

The long-term goal of this project is to improve the sustainability and profitability of the U.S. Christmas tree industry by developing and using genomic tools to produce low cost and high quality Christmas trees with properties desired by consumers. This project is funded by the Specialty Crop Research Initiative Grant 2012-51181-19940 of the USDA’s National Institute of Food and Agriculture. We are currently collaborating with the Christmas Tree Genetic CTG) program at NC State and Plant Computational Biology Lab at University of Connecticut.
Specifically, we will:
- Identify single nucleotide polymorphic markers (SNPs) with predictive power for desirable traits in true firs through a two-step process: Identify candidate genes by RNA sequencing (RNA Seq) and screen SNPs in candidate genes for association with phenotypes using targeted sequencing of genomic DNA.
- Use SNP markers to accelerate production of trees with superior postharvest needle retention and resistance to Phytophthora root rot, and leverage marker-informed breeding with data from a separate industry-supported study of adaptability to climate change.
- Conduct surveys and focus groups to increase our understanding of consumer Christmas tree preferences and improve growers’ ability to respond.
- Educate and advise growers in genetic improvement techniques, seed orchard establishment, and using improved genotypes to produce high quality Christmas trees desired by consumers.
Identification of key molecular markers for needle retention

Needle retention is the capacity to maintain needles set on branches and is associated with needle abscission. By design, trees lose different organs through their lives and this process is called abscission. Most conifers have leaves with a needle-like appearance and therefore, we will refer to needle abscission. Needle abscission occurs in the NAZs (Needle Abscission Zones) and is an active process that involves a complex gene network control. The process involved the degradation of the lamella, which is the “glue” that maintains together the leaf to the tree by enzymatic activity. Then, cell expansion and changes in the turgor pressure allow the separation of the tissue that needs to be removed and finally the set of a protection area which protects the tree from the environment.
Surveys conducted by (NCTA) consistently indicate that “messiness” is arguably the most important issue facing the sustainability of the real Christmas tree industry in its competition with artificial trees (National Christmas Tree Association, 2011). Research and experience has demonstrated that fir species vary considerably in needle holding ability. Based on physiological data from detached branch technique, we have found that much of the within species is under strong genetic control.
The main goal of this project is improving post harvest needle retention using new genomics tools for the identification of gene networks and molecular markers that control this trait in Fraser fir and other Christmas Tree species, and increase the Christmas tree industry’s competitiveness with artificial trees.
Past Projects
Histological Characterization of Fraser Fir Abscission Zone
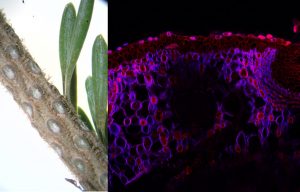
Most conifers have the capacity to retain needles for several years losing them gradually in response to cold acclimation, a natural process referred to as needle abscission (NA). This process only involves a few layers of cells in predictable positions known as abscission zones (AZs).
NA is sometimes severely affected once a tree is cut, shipped and displayed in living rooms, common practices that North American Christmas tree growers and consumers know very well. Christmas tree growers recognize post-harvest needle abscission (PNA) as a common problem but changes in weather patterns and earlier harvest practices are increasing this phenomenon and highlight the need to identify trees with better adaptation.
There is a growing demand for breeding Christmas tree varieties that hold their needles for longer periods of time to encourage more consumers to buy real Christmas trees instead of artificial trees. At the same time, there is a lack of methods that allow us to predict PNA as well as a limited knowledge of the histology and gene regulatory network that controls this important trait.
We designed a system for the histological characterization of AZs in Fraser fir to complement our transcriptome data analysis to find putative control genes that play crucial roles during NA and PNA and could be used as molecular markers. The histological system included fixation, staining and sectioning of AZs collected in the field and at different time points during indoor display followed by examination under bright-field and confocal microscopy. Using these approaches, we described AZ development, identified potential anatomical differences between trees that retain their needles better and will use this information to link physiological and transcriptome data in Fraser fir and other fir species.
This project was possible thanks to the generous support of the Provost’s Professional Experience Program (PEP), and the collaboration with Marcela Rojas-Pierce’s lab and the Cellular and Molecular Imaging Facility (CMIF) at NCSU.
Studies of Fraser Fir Seed Chalcid Infestation
The Christmas tree industry is mainly supplied with seeds from natural stands and genetically improved germplasm from clonal seed orchards (CSOs). Chalcid wasps (Megastigmus spp.) have a negative effect on the value of seedlots by reducing the quantity of viable seeds and represent a risk for national and international seed trade. Seed radiographies, field observations and seed-dissections from 25 Fraser fir (Abies fraseri (Pursh) Poir.) clones were used to determine chalcid infestation in a CSO of the southern Appalachians. We provided direct evidence that Mesopolobus cf. pinus is an ectoparasitoid of M. specularis, proposed a model that linked different aspects of the biology of these species and discussed host–insect phenological relationships. Infestation of seeds in some clones across surveys, suggested a potential genetic component to chalcid wasp’s prevalence. Cold and dark rearing regimens were used to identify changes in adult insect emergence after variation of overwinter conditions. Both the frequency and the time of emerging M. specularis adults from seeds were affected by the duration of the treatments but not statistical differences were found for Mesopolobus cf. pinus. We present evidence for the efficiency of a gravity-seed cleaning method eliminating upwards of 95% of chalcid-infested seeds.
Male M. specularis isolated from Fraser fir seeds in our lab. Photo property of Matthew Bertone and the NC State Plant Disease and Insect Clinic
Female isolated from Fraser fir seeds in our lab Photo property of Matthew Bertone and the NC State Plant Disease and Insect Clinic
For more information consult:
A simple laboratory rearing method for chalcid wasps.dx.doi.org/10.17504/protocols.io.bb3riqm6
Somatic Embryogenesis of Fir species
Fraser fir Christmas trees are one of North Carolina’s largest specialty crops, producing revenue of over $100 million annually. Genetic improvement efforts are underway using traditional plant breeding methods to increase growth, quality, pest resistance, and post-harvest needle retention. However, progress is slow due to intrinsic challenges of breeding a coniferous species with a long generation cycle (8-15 years).
Somatic embryogenesis is a tissue culture cloning technique that generates genetically identical individuals. It is an important biotechnology tools and a key step in the development of genetic transformation and propagation protocols to produce genetically engineered planting stock in the future.
The goal of this project is to improve somatic embryogenesis methods for Fraser fir and thus, provide a pathway for large-scale clonal propagation and genetic engineering in the future.
In conifers, the preferred starting material for somatic embryogenesis is immature embryos extracted from immature cones during late June.
Fraser fir cultured as Christmas trees is one of North Carolina’s largest specialty crops with an annual revenue exceeding $100 million. The most severe disease in Fraser fir nurseries and plantations is Phytophthora root rot which afflicts the industry with an estimated $6 million in losses annually. Because Fraser fir has no innate resistance, the NCSU Christmas Tree Genetics Program has begun evaluation of two exotic species that show promise as quality Christmas tree species and also possess some Phytophthora resistance.
Genetic control of resistance to a single Phytophthora genotype has been confirmed. The research will assess resistance in these fir species to a broader array of Phytophthora genotypes and use next generation sequencing technology to develop DNA markers to select for resistance in genetic field trials.
The technology and knowledge generated will be applied toward developing Phytophthora-resistant fir planting stock targeted for sites with known disease problems and will ultimately abate the adverse impact of this disease on the state’s Christmas tree industry.
A survey of Phytophthora species in Fraser fir Christmas tree plantations in the southern Appalachians
Martin Pettersson conducted a survey of Phytophthora in Fraser fir Christmas tree plantations. Symptomatic Fraser fir roots have been collected in commercial production fields distributed over 14 counties in three states in the Southern Appalachians. Roots from individual trees were transferred to a selective media and small tufts of mycelium were taken from the pure cultures that morphologically Phytophthora. After DNA extraction, at least six species were identified.
For more information consult:
Identification of Signal Molecules that Play a Role in Disease Resistance (edit)
Be able to identify plant traits that are correlated with disease resistance is an interesting approach and offer a physiological tool for breeding programs.
Through the use of the mass spectrometry we have identified certain chemical compounds that seems routinely to be in much higher concentrations in two different firs, one of them with significance Balsam Woolly Adelgid (BWA) resistance. (use pictures from the website)
Ethan Bucholz, a mater student in our lab, is attempting to alter the concentration of those components in the headspace of closed environments, to assess its effect on BWA health and reproductive success. Bucholz will also compare (solid face microextraction) analyses across a wide range of susceptibility within the Abies genus to identify potential differences and similarities between the various forms of susceptibility (from risk to resistant).