Research II
The ability to use DNA markers to identify parentage could eliminate the cost associated with control-pollinations in tree improvement programs, a concept referred to in the literature as “breeding without breeding”. Parental analyses will be carried out to attempt to identify the paternal parent of each progeny. The results will be used to determine the level of panmixis in the seed orchard and the usefulness of this approach to reduce time and costs of our Fraser fir tree improvement efforts.
The ability to use DNA markers to identify parentage could eliminate the cost associated with control-pollinations in tree improvement programs, a concept referred to in the literature as “breeding without breeding”. Parental analyses will be carried out to attempt to identify the paternal parent of each progeny. The results will be used to determine the level of panmixis in the seed orchard and the usefulness of this approach to reduce time and costs of our Fraser fir tree improvement efforts.
Projects in the lab are cross-disciplinary providing a nourishing research environment to undergraduate and graduate students, and postdoctoral researchers. Our studies are focused on answering scientific questions, offering a link between academia and industry and translating our findings into solutions for tree growers and stakeholders. Our main interests are conifers and we are working hard to consolidate a network between different universities and industry, specially the Christmas Tree Industry in North Carolina.
Genomics-based Technologies in Forest Tree Breeding
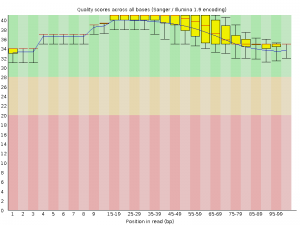
Biotechnology and bioinformatics offer a wide spectrum of possibilities to decode, study and propose models based on tree genome sequences. The improvement and application of new molecular and genetic tools could sharply reduce the time required to grow the trees and help to identify trees with certain characteristics that confer resistance to pest infestation and environmental changes.
Development and use of genomic tools to improve firs for use as Christmas trees

The long-term goal of this project is to improve the sustainability and profitability of the U.S. Christmas tree industry by developing and using genomic tools to produce low cost and high quality Christmas trees with properties desired by consumers.
Specifically, we will:
- Identify single nucleotide polymorphic markers (SNPs) with predictive power for desirable traits in true firs through a two-step process: Identify candidate genes by RNA sequencing (RNA Seq) and screen SNPs in candidate genes for association with phenotypes using targeted sequencing of genomic DNA.
- Use SNP markers to accelerate production of trees with superior postharvest needle retention and resistance to Phytophthora root rot, and leverage marker-informed breeding with data from a separate industry-supported study of adaptability to climate change.
- Conduct surveys and focus groups to increase our understanding of consumer Christmas tree preferences and improve growers’ ability to respond.
- Educate and advise growers in genetic improvement techniques, seed orchard establishment, and using improved genotypes to produce high quality Christmas trees desired by consumers.

Identification of key molecular markers for needle retention in firs
Needle retention is the capacity to maintain needles set on branches and is associated to needle abscission. By design, trees loose different organs through their lives and this process is called abscission. Most conifers have leaves with a needle-like appearance and therefore, we will refer to needle abscission. Needle abscission occurs in the NAZs (Needle Abscission Zones) and is an active process that involved a complex gene network control. The process involved the degradation of the lamella, which is the “glue” that maintains together the leaf to the tree by enzymatic activity. Then, cell expansion and changes in the turgor pressure allow the separation of the tissue that need to be removed and finally the set of a protection area which protect the tree from the environment.
Surveys conducted by (NCTA) consistently indicate that ‘messiness’ is arguably the most important issue facing the sustainability of the real Christmas tree industry in its competition with artificial trees (National Christmas Tree Association 2011). Research and experience has demonstrated that fir species vary considerably in needle holding ability. Based on physiological data from detached branch technique, we have found that much of the within species is under strong genetic control.
The main goal of this project is improving post harvest needle retention using new genomics tools for the identification of gene networks and molecular markers that control this trait in Fraser fir and other Christmas Tree species, and increase the Christmas tree industry’s competitiveness with artificial trees.
Developing Phytophthora-resistant fir Christmas trees
Fraser fir cultured as Christmas trees is one of North Carolina’s largest specialty crops with an annual revenue exceeding $100 million. The most severe disease in Fraser fir nurseries and plantations is Phytophthora root rot which afflicts the industry with an estimated $6 million in losses annually. Because Fraser fir has no innate resistance, the NCSU Christmas Tree Genetics Program has begun evaluation of two exotic species that show promise as quality Christmas tree species and also possess some Phytophthora resistance.
Genetic control of resistance to a single Phytophthora genotype has been confirmed. The research will assess resistance in these fir species to a broader array of Phytophthora genotypes and use next generation sequencing technology to develop DNA markers to select for resistance in genetic field trials.
The technology and knowledge generated will be applied toward developing Phytophthora-resistant fir planting stock targeted for sites with known disease problems and will ultimately abate the adverse impact of this disease on the state’s Christmas tree industry.

Breeding without breeding
The ability to use DNA markers to identify parentage could eliminate the cost associated with control-pollinations in tree improvement programs, a concept referred to in the literature as “breeding without breeding”. Parental analyses will be carried out to attempt to identify the paternal parent of each progeny. The results will be used to determine the level of panmixis in the seed orchard and the usefulness of this approach to reduce time and costs of our Fraser fir tree improvement efforts.